Understanding the Number Of Protein Domains is central to annotating proteins, interpreting function, and comparing genomes. This article explores common pitfalls in estimating the Number Of Protein Domains, and how to avoid them to improve accuracy and reproducibility.
Common Pitfalls In Estimating The Number Of Protein Domains
Key Points
- Inconsistent domain definitions across databases can skew counts and complicate comparisons.
- Boundary ambiguity between adjacent domains can inflate or deflate estimates depending on detection thresholds.
- Choice of detection method (e.g., HMM-based vs. motif-based) heavily influences the Number Of Protein Domains reported.
- Partial sequences, truncations, or isoforms may reveal only fragments of domains, leading to underestimation.
- High redundancy or multi-domain architectures require careful segmentation and context-aware interpretation to avoid double counting.
Definition and scope of a domain
Clarify what constitutes a domain in the protein you are analyzing and what you mean by the Number Of Protein Domains. Some definitions focus on structure, others on sequence motifs or functional units. Align your approach with the question you want to answer and with the standards used in your field to minimize surprises when comparing results. When you publish, specify which domain model libraries and length criteria you used so others can reproduce your counts.
Boundary detection and segmentation challenges
Most domain prediction tools attempt to segment a sequence into regions that match known domain models. However, domain boundaries are often fuzzy, especially in linker regions or in proteins with novel architectures. Small shifts in boundaries can change the Number Of Protein Domains by one or more units, affecting downstream analyses. Consider reporting boundary uncertainty and using consensus annotations from multiple tools to stabilize counts.
Method choice and parameter sensitivity
Different software packages apply different criteria for domain hits, such as coverage thresholds, e-value cutoffs, or domain model libraries. When you change methods or parameters, you may observe substantial swings in the estimated count, so document the settings and consider multi-method consensus. If possible, benchmark your pipeline against curated datasets to understand method-specific biases.
Impact of sequence quality and isoforms
Draft genomes, low-coverage sequencing, or alternative splicing can produce truncated or fragmentary sequences. These artifacts may hide whole domains or present only partial matches, biasing the Number Of Protein Domains upward or downward. Use curated, high-quality sequences when possible, and report the version of the transcript or protein used. When dealing with isoforms, consider whether to count each isoform separately or to aggregate by gene context.
Evolutionary and architectural context
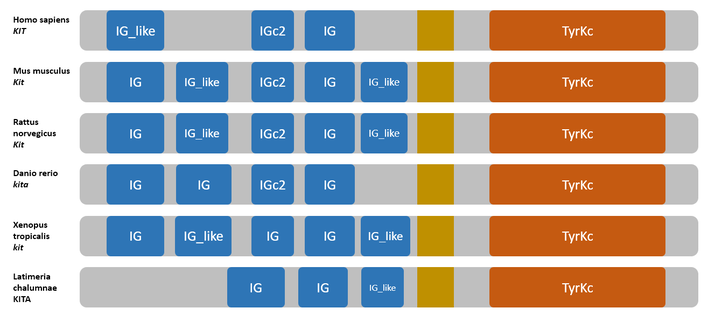
Organisms with frequent domain shuffling, duplications, or domain losses may display atypical architectures. A straightforward count may miss functional nuance, so pair domain counts with structural or functional annotations to avoid misinterpretation. In some clades, lineage-specific expansions can dramatically alter the observed Number Of Protein Domains, which is a sign to interpret counts in a broader evolutionary frame.
Best practices to improve accuracy
Adopt a transparent workflow: define what counts as a domain, choose a consistent set of domain models, validate with manual inspection where feasible, and report uncertainty. When comparing studies, align the definitions and methods to interpret differences correctly. Document how you define the Number Of Protein Domains and provide enough detail for someone else to reproduce your results.
What defines a domain for analysis in practice?
+In practice, defining a domain often blends structure, sequence motifs, and function. A practical approach is to specify a minimum domain length, a required coverage of the model, and whether partial matches count. Document the criteria you apply and ensure they are consistently used across all sequences to derive the Number Of Protein Domains.
Why do different tools give different domain counts for the same protein?
+Different tools use distinct domain libraries, scoring schemes, and thresholds. HMM-based methods may emphasize domain presence differently than motif-based approaches, and some tools allow partial matches that others do not. Running multiple methods and comparing results helps reveal method-specific biases and guides a more robust conclusion about the Number Of Protein Domains.
How can partial sequences affect domain counting?
+Partial sequences from fragments or isoforms can obscure full-domain boundaries, leading to undercounting, or can produce misleading partial hits that inflate the count. To minimize impact, use complete or well-annotated isoforms when possible and explicitly state how partial data were handled in your analysis.
What steps reduce uncertainty in domain counts?
+Adopt a defined and documented counting rule, use a consensus approach across multiple domain models, validate surprising results with manual inspection, and report uncertainty ranges. When possible, benchmark your pipeline against curated reference datasets, and clearly describe parameter choices and and the domain model libraries used to derive the Number Of Protein Domains.