Mammalian Orthoreovirus Σns is a non-structural RNA-binding protein that modulates RNA metabolism during infection, influencing how the virus replicates and interacts with the host cell. This article examines its structure and role in infection, highlighting how Σns contributes to replication efficiency, genome flexibility, and host-pathogen interactions.
Key Points
- Σns binds viral RNA and helps assemble replication and transcription complexes.
- The RNA-binding surface and potential dimerization support efficient genome handling.
- Localization to viral factories concentrates replication machinery and protects RNA.
- Σns interacts with host pathways to influence innate immune signaling.
- Structural features of Σns may be targeted by antiviral strategies.
Structure of Mammalian Orthoreovirus Σns

The Σns protein is relatively small but functionally rich. It features an RNA-binding domain that recognizes single- and double-stranded RNA motifs and a flexible linker that may facilitate interactions with other viral or host factors. Bioinformatic predictions suggest an alpha-helical core with regions capable of dimerization, supporting cooperative RNA binding. In infected cells, Mammalian Orthoreovirus Σns localizes to cytoplasmic compartments associated with replication factories, where it can coordinate RNA processing with other viral proteins.
Domain organization
The N-terminal region contains the RNA-binding surface, followed by a central linker that allows conformational changes. A predicted oligomerization interface helps Σns form functional assemblies that stabilize RNA substrates during replication and transcription.
Interactions and localization
Σns interacts with viral core proteins and host factors involved in RNA metabolism. Its localization to viral factories suggests a role in concentrating replication machinery and shielding viral RNA from host defenses.
Role of Mammalian Orthoreovirus Σns in Infection
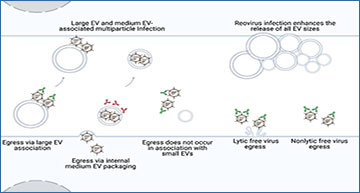
During infection, Mammalian Orthoreovirus Σns contributes to efficient genome replication and transcription by supporting RNA handling and complex assembly. It acts as an RNA chaperone, helping to unfold or rearrange RNA structures to permit efficient transcription by the viral polymerase complex. The protein also modulates host responses, potentially dampening innate signaling pathways to favor viral gene expression. As infection progresses, Σns dynamics correlate with shifts in replication factory activity, aligning with the need for rapid genome production and virion assembly.
Genome replication and transcription
In core replication complexes, Σns binds viral RNA templates, stabilizes intermediate structures, and assists in packaging signals for transcription. Its activity is coordinated with other reovirus non-structural proteins to optimize strand separation and RNA synthesis.
Host interactions and immune response
Σns can influence host RNA-binding networks and innate immune sensors. By modulating RNA accessibility and localization, it may reduce detection by pattern recognition receptors, giving the virus a window to replicate before antiviral defenses are fully engaged.
Temporal expression and regulation
Expression of Mammalian Orthoreovirus Σns is tightly timed during the early to mid stages of infection, ensuring that RNA metabolism is aligned with replication factory maturation and genome packaging needs.
What is the function of Mammalian Orthoreovirus Σns in infection?
+Mammalian Orthoreovirus Σns is a non-structural RNA-binding protein that supports RNA metabolism during infection, aiding replication, transcription, and the coordination of replication factories.
How does Σns interact with host cells?
+Σns engages with host RNA-binding factors and signaling pathways, potentially modulating antiviral responses to favor viral gene expression.
Where is Mammalian Orthoreovirus Σns localized during infection?
+Σns localizes to cytoplasmic replication factories and associated RNA-processing sites, facilitating coordination of viral RNA synthesis.
Can targeting Σns disrupt reovirus replication?
+Disrupting the RNA-binding surface or oligomerization interface of Σns could hamper RNA metabolism and replication, representing a theoretical antiviral strategy needing further research.
What structural features of Σns make it a potential antiviral target?
+Key features include the RNA-binding domain, dimerization interfaces, and flexible linkers that coordinate interactions with viral and host factors.